Open Science and the development of open-access tools is an increasingly popular way to make science accessible, reproducible, scalable, and replicable (Lowndes et al., 2017).
The most popular open-access platform for analysis and reporting of data is R (R Development Core Team, 2016). R provides more than 5000 packages for data exploration, visualisation, modelling and statistical analysis.
In addition, the R Shiny and RMarkdown packages allow for the creation of web tools, allowing users to interact with science outputs and methods, without any requirement for software (only web access is required) or a need to understand complex code. Another advantage of R Shiny is that it allows scientists to directly create web tools without any of the disadvantages associated with using 3rd party programmers (e.g. high cost, difficulty communicating ideas or interpreting science).
RMarkdown is used to document code used within RShiny web apps, supporting reproducibility, transparency and reuse of existing code.
Within Cefas (Lowestoft and Weymouth) and collaboration with University of East Anglia, multiple tools have been created and more are in development .
These tools are designed to be open access and shared to drive scientific innovation . To ensure Cefas tools/applications are developed and deployed in a way which satisfies Open Science criteria, a framework (Fig. 1) has been developed. This framework ensures that tools are based on solid foundations of:
- access to data using Cefas systems,
- high quality, open access peer review publications, and transparency of methods using code repositories such as gitHub.
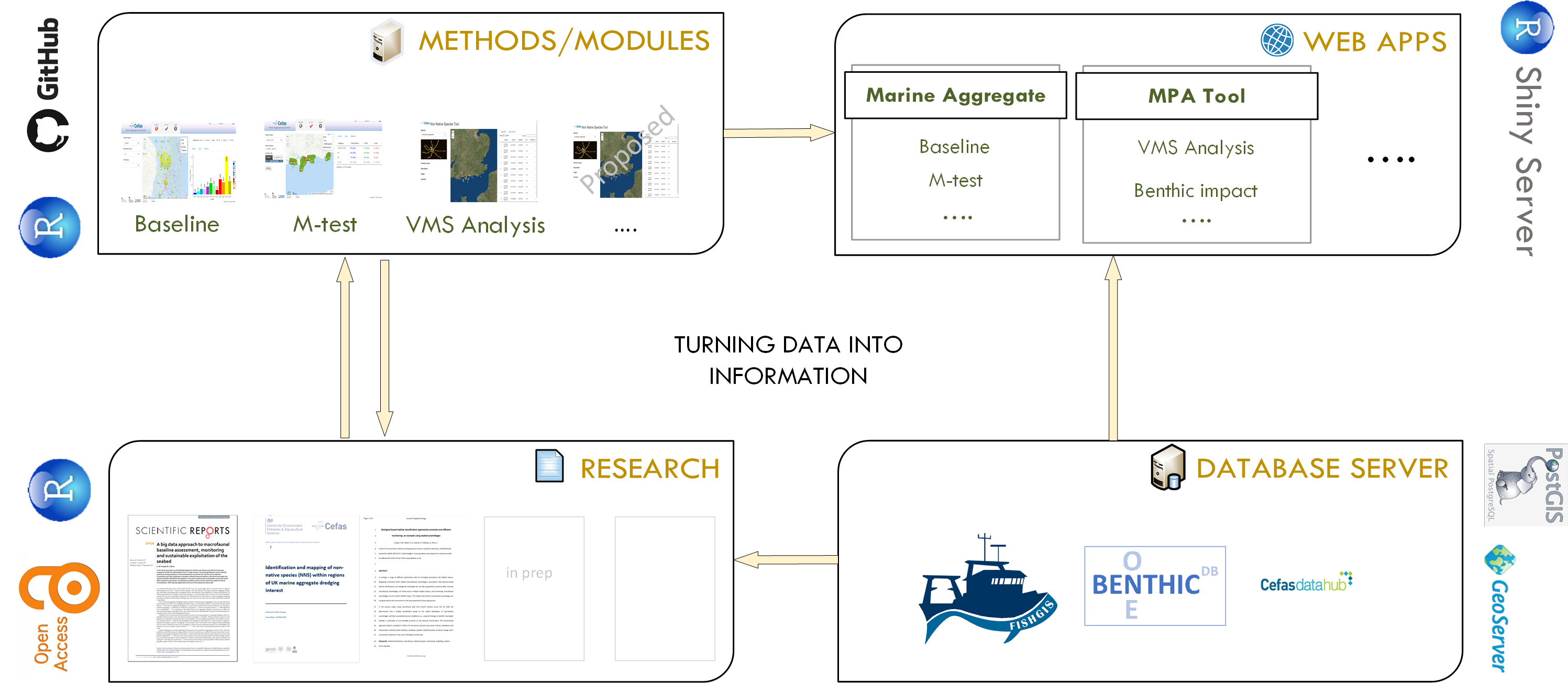 Fig.1 - Open Science Framework workflow
Fig.1 - Open Science Framework workflow